1. HBV-TCR T cell therapy
We have been the first lab to show that T cell receptor-engineered T cells specific for HBV antigens can target HBV-related HCC tumors in patients with primary or secondary HCC(1). Our approach leverages on the flexibility of mRNA technology to produce HBV-specific T cell receptors engineered T cells that are infused in escalating doses in patients(2, 3). Such approach avoids genetic manipulation of the adoptively transfer T cells and the transient expression of TCR reduces their on-target off-tumor lysis of HBV infected hepatocytes. We are now developing novel methods to develop an allogenic TCR-redirected T cell treatment approach that can avoid host versus graft (HvG) reaction, and to increase the antiviral activity of engineered T cells for antiviral use (HBV and SARS-CoV-2 infection).
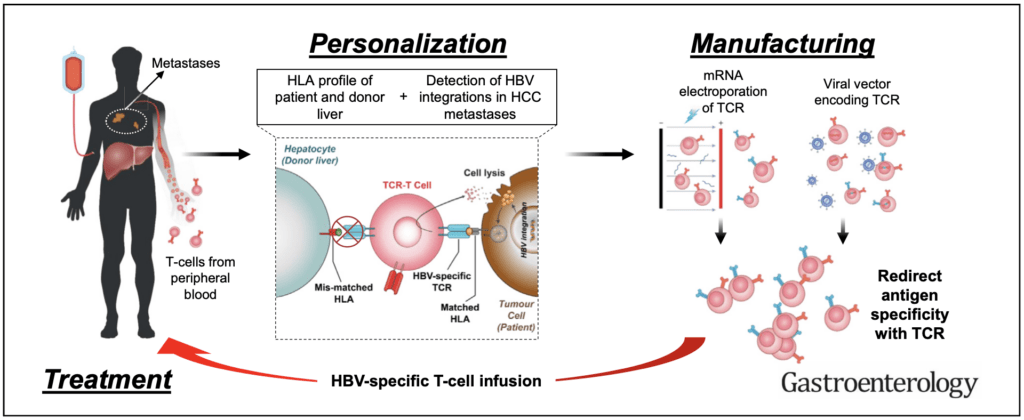
2. T cell immune response in Hepatitis B Virus (HBV) infection
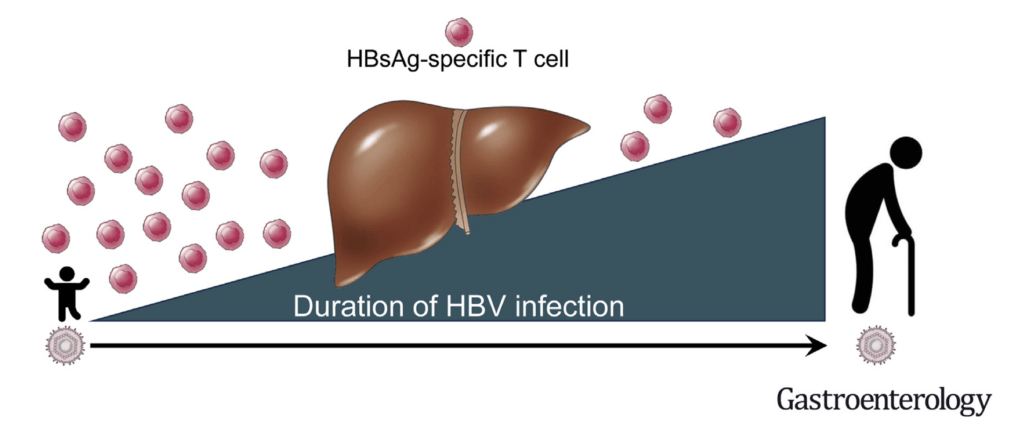
We aim to integrate HBV-specific T cell immune profiles with the classical virological (HBsAg, HBeAg) and biochemical (ALT) characterization of patients with chronic HBV infection. We are developing a robust and rapid whole blood assay (WBA)(4) that allows the measurement of the functional secretome of HBV-specific T cells in large patient populations. Machine learning algorithms separate CHB patients within similar clinical phases into clusters with distinct HBV-specific immune profiles. An AI-guided classification of patients within similar clinical disease phases based on their antiviral T cell function will provide a novel way for interpreting host-viral interactions and signpost the selection of novel immunotherapies.
3. T cells in SARS-CoV-2 infection and vaccination.
After the start of the COVID-19 pandemic, we have been actively involved in the characterization of SARS-COV2 specific T cell immune response in COVID-19 and SARS convalescent and healthy individuals. We were among the first lab to profile T cells in SARS-CoV-2 infected individuals (6), and to demonstrate that the early appearance of functional SARS-CoV-2 -specific T cells correlated with rapid viral control in patients (7,8). We also showed that SARS-CoV-2 specific T cells are highly efficient particularly in asymptomatic SARS-CoV-2 infection (9), data that support the importance of T cells in protection from severe COVID-19. In addition, we were involved in studies characterizing the temporal association of vaccine-induced T cells appearance and reduction of symptomatic infection (10), in the comparative analysis of T cells induced by inactivated virus vaccines (11) and on the development of methods to rapidly evaluate SARS-CoV-2 T cells (5,12). Since the initial site of SARS-CoV-2 infection are the upper airways, we are now focusing our work on the analysis of T cell localized at the site of primary infection. Initial findings in individuals with breakthrough infection showed that this tissue-resident CD8 T cell population appears to be induced only by infection and by not parenteral vaccination (13).

4. Other viruses
Hematopoietic stem cell transplant (HSCT) recipients who received immunosuppressive treatments for graft preservation frequently experience reactivation or infection with human cytomegalovirus (hCMV), which can lead to significant disseminated pathologies. Even though hCMV-specific T cells and NK cells are important for the control of hCMV infection/reactivation, existing methods and commercial assays of hCMV-specific T cell and NK cell analysis requires complex isolation and culture processes, making such assays unfeasible for large-scale use in the clinical setting.
Hence, we adapted the rapid whole blood assay to easily measure the adaptive (hCMV-specific T cell) and innate (NK cell) immune response against hCMV in the whole blood of individuals. This assay will then be used to define the hCMV cellular immunity in different groups of HSCT recipients and validate whether such immunological information can guide the clinical management of post-transplant hCMV reactivation/infection.
References
- Qasim, W., Brunetto, M., Gehring, A. J., Xue, S. A., Schurich, A., Khakpoor, A., . . . Bertoletti, A. (2015). Immunotherapy of HCC metastases with autologous T cell receptor redirected T cells, targeting HBsAg in a liver transplant patient. Journal of Hepatology, 62(2), 486-491.
- Kah, J., Koh, S., Volz, T., Ceccarello, E., Allweiss, L., Lütgehetmann, M., . . . Protzer, U. (2017). Lymphocytes transiently expressing virus-specific T cell receptors reduce hepatitis B virus infection. Journal of Clinical Investigation, 127(9), 3177-3188.
- Tan, A. T., Yang, N., Lee Krishnamoorthy, T., Oei, V., Chua, A., Zhao, X., Tan, H. S., Chia, A., Le Bert, N., Low, D., Tan, H. K., Kumar, R., Irani, F. G., Ho, Z. Z., Zhang, Q., Guccione, E., Wai, L. E., Koh, S., Hwang, W., Chow, W. C., & Bertoletti, A. (2019). Use of expression profiles of HBV-DNA integrated into genomes of hepatocellular carcinoma cells to select T cells for immunotherapy. Gastroenterology, 156(6), 1862-1876.e9. doi:10.1053/j.gastro.2019.01.251
- Tan AT, Lim JME, Le Bert N, Kunasegaran K, Chia A, Qui MDC, Tan N, Chia WN, de Alwis R, Ying D, Sim JXY, Ooi EE, Wang L-F, Chen MIC, Young BE, Hsu LY, Low JGH, Lye DC, Bertoletti A. Rapid measurement of SARS-CoV-2 spike T cells in whole blood from vaccinated and naturally infected individuals. J Clin Invest. 2021 Sep 1;131(17):e152379. doi: 10.1172/JCI152379.
- Le Bert N, Gill US, Hong M, Kunasegaran K, Tan DZM, Ahmad R, Cheng Y, Dutertre CA, Heinecke A, Rivino L, Tan A, Hansi NK, Zhang M, Xi S, Chong Y, Pflanz S, Newell EW, Kennedy PTF, Bertoletti A. Effects of Hepatitis B Surface Antigen on Virus-Specific and Global T Cells in Patients With Chronic Hepatitis B Virus infection. Gastroenterology. 2020 Aug;159(2):652-664. doi: 10.1053/j.gastro.2020.04.019.
- Le Bert N, Tan AT, Kunasegaran K, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457-462.
- Tan AT, Yang N, Lee Krishnamoorthy T, Oei V, Chua A, Zhao X, Tan HS, Chia A, Le Bert N, Low D, Tan HK, Kumar R, Irani FG, Ho ZZ, Zhang Q, Guccione E, Wai LE, Koh S, Hwang W, Chow WC, Bertoletti A. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Reports. 2021 Feb 9;34:108728. doi: 10.1016/j.celrep.2021.108728.
- Swadling L, Diniz MO, Schmidt NM, et al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature. 2022;601:110-117. doi:10.1038/s41586-021-04186-8
- Le Bert N, Clapham HE, Tan AT, Chia WN, Tham CYL, Lim JM, Kunasegaran K, Tan LWL, Dutertre CA, Shankar N, Lim JME, Sun LJ, Zahari M, Tun ZM, Kumar V, Lim BL, Lim SH, Chia A, Tan YJ, Tambyah PA, Kalimuddin S, Lye D, Low JGH, Wang LF, Wan WY, Hsu LY, Bertoletti A, Tam CC. Highly functional virus-specific cellular immune response in asymptomatic SARS-CoV-2 infection. Journal of Experimental Medicine. 2021 May 3;218(5):e20202617. doi: 10.1084/jem.20202617. PMID: 33646265; PMCID: PMC7927662.
- Kalimuddin S, Tham CYL, Qui M, de Alwis R, Sim JXY, Lim JME, Tan HC, Syenina A, Zhang SL, Le Bert N, Tan AT, Leong YS, Yee JX, Ong EZ, Ooi EE, Bertoletti A, Low JG. Early T cell and binding antibody responses are associated with COVID-19 RNA vaccine efficacy onset. Med. 2021 Jun 11;2(6):682-688.e4. doi: 10.1016/j.medj.2021.04.003. Epub 2021 Apr 8. PMID: 33851143; PMCID: PMC8030737.
- Lim JME, Hang SK, Hariharaputran S, Chia A, Tan N, Lee ES, Chng E, Lim PL, Young BE, Lye DC, Le Bert N, Bertoletti A, Tan AT. A comparative characterization of SARS-CoV-2-specific T cells induced by mRNA or inactive virus COVID-19 vaccines. Cell Reports Medicine. 2022 Nov 15;3(11):100793. doi: 10.1016/j.xcrm.2022.100793. Epub 2022 Oct 6. PMID: 36257326; PMCID: PMC9534788.
- Schwarz M, Torre D, Lozano-Ojalvo D, et al. Rapid, scalable assessment of SARS-CoV-2 cellular immunity by whole-blood PCR. Nat Biotechnol. 2022;40:1680-1689. doi:10.1038/s41587-022-01347-6
- Lim JME, Tan AT, Le Bert N, Hang SK, Low JGH, Bertoletti A. SARS-CoV-2 breakthrough infection in vaccinees induces virus-specific nasal-resident CD8+ and CD4+ T cells of broad specificity. Journal of Experimental Medicine. 2022 Oct 3;219(10):e20220780. doi: 10.1084/jem.20220780.
